Structural biology consultant on Kolabtree Jennifer Huen writes about how scientists worldwide are working to find solutions to the COVID-19 pandemic.
As I write these words, medical staff around the world are working at full capacity to treat infected patients, scientists are hurriedly conducting experiments to develop a suitable COVID-19 vaccine, and volunteers are delivering food and supplies to vulnerable people. These are only a handful of the numerous selfless acts people are undertaking to assist everyone around them.
Our collective experience during the SARS epidemic in 2002-2003 has led to swift actions and behavior changes by the government and general population in order to contain viral spread and accelerate urgent research [1]. Already, over a thousand primary and review articles have been published on SARS-CoV2 since its first appearance in the public eye in December of last year. Lab suppliers are streamlining orders for COVID-19 related reagents and media outlets are providing free access to COVID-19 articles, such as those from Science magazine (https://www.sciencemag.org/). The Protein Data Bank, a resource for high resolution molecular structures solved by the scientific community, has seen over a hundred protein structures deposited, the first at the beginning of February [2]. Such research as well as past work on coronaviruses have led to the expedition of clinical trials worldwide [3-5].
With the bounty of data made freely available, I generated the image below to represent a snapshot of how SARS-CoV2 infects its host. The protein molecules illustrated in this image were obtained from molecular structure data provided by several research groups [6-8]. One of these groups has provided structures of all SARS-CoV2 proteins and their interactions with human protein targets (http://korkinlab.org/wuhan).
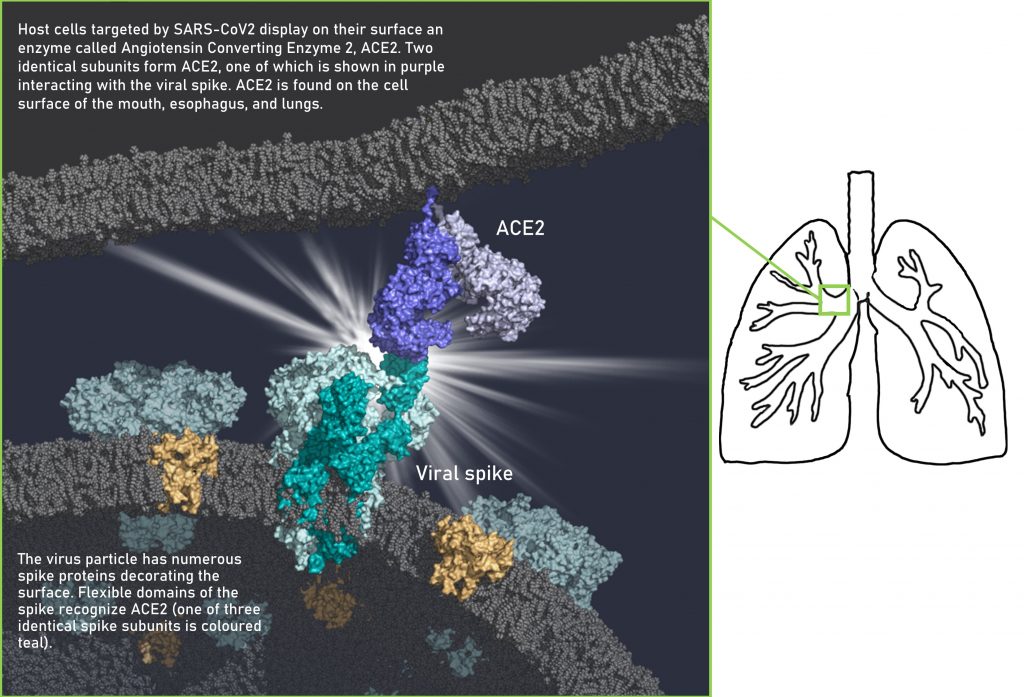
During SARS-CoV infection, host cells either engulf the virus particle or the virus membrane directly fuses to the host membrane [9,10]. These two actions are first mediated by the viral spike protein binding the enzyme displayed on the host surface, Angiotensin Converting enzyme 2, ACE2. Scientists have shown since February that ACE2 is the enzyme targeted by SARS-CoV2 [11-14]. Once the virus spike binds to ACE2, host enzymes called proteases cut a specific region of the viral spike, allowing the spike to rearrange itself. This action makes way for part of the spike to insert into the host membrane, allowing the virus membrane to fuse to it and extrude its genome inside the host [15,16]. The virus can also enter its host by becoming engulfed into vesicles, where it can then deposit its genome inside the host [17,12]. By interfering with any of the steps for SARS-CoV2 entry, infection can be halted. Molecular structures of viral proteins play an important role in drug research as its akin to having a map of unknown territory, revealing key regions on the target molecule.
This work is only one of hundreds of articles that were expedited for publication by science journals, by-passing the peer-review process, in order to reach the public faster. The amount of progress we’ve made in finding solutions to COVID-19 could only have been possible with the help from the many resourceful and innovative people worldwide. To see how you can contribute, please see the list of opportunities below.
I have always appreciated the elegant structures and functions of biological molecules. Now, I’m greatly appreciative of the speed in which scientists and the general public worldwide are dedicating their energies toward a common goal. It will be our collaborative efforts that will pull us through this pandemic.
Links for volunteering, innovation, and donation resources:
Volunteering activities may involve helping with COVID-19 diagnoses, fact-checking and addressing misinformation, delivery of medical reagents or donations of personal protective equipment. This list is not comprehensive and contains volunteer opportunities mostly around the Toronto, Canada area.
- https://covid19resources.ca/ (scientists needed for diagnostics and other lab/non-lab related tasks around Toronto, Canada)
- https://crowdfightcovid19.org/volunteers (researchers to assist in COVID-19 research projects such as transcribing data and bioinformatics)
- https://www.helpfulengineering.org/volunteer/ (engineers to assist in various technical/non-technical tasks related to COVID-19)
- https://www.kaggle.com/allen-institute-for-ai/CORD-19-research-challenge (AI experts to contribute data mining tools and other expertise, awards available)
- https://www.coventchallenge.com/ (technical experts to design ventilators with help creating prototypes)
- https://helpwithcovid.com/ (search hub connecting volunteers to different covid-19 projects)
- http://uhnopenlab.ca/project/hotline/ (delivery of food and other essentials for vulnerable seniors around Toronto, Canada)
- https://masksfordocs.com/ (donations of protective supplies for medical workers)
- https://www.conquercovid19.ca/ (sourcing medical supplies for Canadian doctors and nurses)
Need to consult an expert for help with researching potential solutions to the COVID-19 pandemic? Get help with coronavirus-related fact-checking, writing and research. Get in touch with qualified scientists on Kolabtree. VIEW CORONAVIRUS EXPERTS
References:
- Threats, I.o.M.U.F.o.M., Learning from SARS: Preparing for the Next Disease Outbreak: Workshop Summary. 2004.
- Jin, Z., et al., Structure of Mpro from COVID-19 virus and discovery of its inhibitors. bioRxiv, 2020: p. 2020.02.26.964882.
- Jiang, S., Don’t rush to deploy COVID-19 vaccines and drugs without sufficient safety guarantees, in Nature. 2020, Springer Nature Limited. p. 321.
- NIH clinical trial of investigational vaccine for COVID-19 begins. 2020, National Institutes of Health.
- WHO Director-General’s opening remarks at the media briefing on COVID-19 – 27 March 2020. 2020, World Health Organization.
- Cui, H., et al., Structural genomics and interactomics of 2019 Wuhan novel coronavirus, 2019-nCoV, indicate evolutionary conserved functional regions of viral proteins. bioRxiv, 2020: p. 2020.02.10.942136.
- Yan, R., et al., Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science, 2020. 367(6485): p. 1444-1448.
- Song, W., et al., Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog, 2018. 14(8): p. e1007236.
- Groneberg, D.A., R. Hilgenfeld, and P. Zabel, Molecular mechanisms of severe acute respiratory syndrome (SARS). Respir Res, 2005. 6: p. 8.
- Yang, Z.Y., et al., pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J Virol, 2004. 78(11): p. 5642-50.
- Zhou, P., et al., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 2020. 579(7798): p. 270-273.
- Ou, X., et al., Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun, 2020. 11(1): p. 1620.
- Xiaowei Li, et al., Molecular immune pathogenesis and diagnosis of COVID-19. Journal of Pharmaceutical Analysis, 2020.
- Meng, T., et al., The insert sequence in SARS-CoV-2 enhances spike protein cleavage by TMPRSS. bioRxiv, 2020: p. 2020.02.08.926006.
- Belouzard, S., V.C. Chu, and G.R. Whittaker, Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci U S A, 2009. 106(14): p. 5871-6.
- Li, F., Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu Rev Virol, 2016. 3(1): p. 237-261.
- Inoue, Y., et al., Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J Virol, 2007. 81(16): p. 8722-9.







